Project 1: The role of Polycomb Group Proteins in Plant Development
Epigenetic memory factor – how do they select target regions?
The Polycomb Group (PcG) pathway creates locally restricted tight chromatin packaging. A PcG-mediated memory is metastable and autonomous at each locus, meaning that each locus in each cell can acquire or lose the PcG-mediated epigenetic memory independently. Our group aims to better understand PcG-dependent gene regulation in plants. We are particularly interested in understanding how PcG proteins choose their target regions and how PcG regulation integrates into plant development.
In the last years, substantial progress has been made in understanding how PcG protein complexes recognize their target genes. Epigenetic regulation is underpinned by a genetic code composed of several distinct cis-elements that recruit sequence-specific transcription factors that act as PcG recruiters. These cis-elements are peppered across several 1000 base-pair long regions and act in a combinatorial way, which modulates the strength of PcG recruitment in different cell types individually for each locus (Figure 1).
Our group has demonstrated that TELOMERE REPEAT BINDING FACTORs (TRBs), which bind to telobox motifs, directly recruit Polycomb Repressive Complex 2 to target regions [1,2]. Polycomb Repressive Complex 2 is a subunit of the PcG pathway. As their name indicates, TRBs were originally described by their propensity to associate with the telomeric repeats at the ends of chromosomes. Telomeric repeats are comprised of direct repeats of several hundred teloboxes. Thus, there could be a competition between telomeres and PcG target genes for the binding of TRBs. In the coming years, we will follow up on the hypothesis that telomere dynamics impact PcG-mediated memory states.
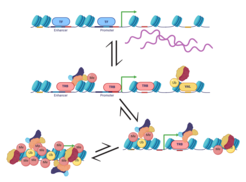
Figure 1: A Schematic representation of the memory path of an expressed gene as it becomes epigenetically repressed. Top, an expressed gene (line) wrapped around nucleosomes (blue) that is predisposed towards stable epigenetic repression by the presence of PcG recruiting cis-motifs (telobox as red line, RY motifs as yellow line). The expressed state is in equilibrium with a non-expressed state (middle), transition between the states is regulated by the availability of activating (TF, blue circle) and repressing (TRBs, red; VALs, yellow) transcription factors (middle). If the non-expressed state persists, PcG-associated telobox- and RY-motifs recruit their binding factors, TRB- and VAL-family proteins, respectively. TRB and VAL-family proteins, in turn, respectively recruit POLYCOMB REPRESSIVE COMPLEXES 2 and 1 (complex pictograms), which carry out histone modifications such as tri-methylation of lysine 27 of histone H3 (red circle, labeled Me) and mono-ubiquitination of lysine 121 of histone H2A (yellow circle, labeled Ub)). These modification and protein-protein interactions between PRCs establish a fully compacted state that is much less reversible (bottom).
[1] Zhou Y, Hartwig B, Velikkakam James G, Schneeberger K, Turck F. (2016) Complementary activities of TELOMERE REPEAT BINDING proteins and Polycomb Group complexes in transcriptional regulation of target genes. Plant Cell 28(1):87-101. doi 10.1105/tpc.15.00787 open access
[2] Zhou, Y., Wang, Y., Krause, K., Yang, T., Dongus, J. A., Zhang, Y., Turck, F. (2018) Telobox motifs recruit CLF/SWN–PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nature Genetics 50(5):638-644, doi: 10.1038/s41588-018-0109-9 open access
