Quantitative approaches to multicellular dynamics in plant development
Throughout plant development, cells grow, divide, and differentiate to form tissues and organs that perform specific functions. This process results from the interplay of cell signalling (mediated by gene regulatory networks and hormone transport) and mechanical cues, in response to environmental signals (e.g., temperature and light). However, we still have a limited quantitative understanding of the complex multicellular dynamics that occur in developing plant tissues, as well as how these dynamics lead to reproducible developmental outcomes.
In our group at the MPIPZ, we study the multicellular dynamics of plant developmental processes by combining confocal microscopy imaging of developing plant tissues, quantitative image analysis, and mathematical modelling.
Dynamics of cellular patterning
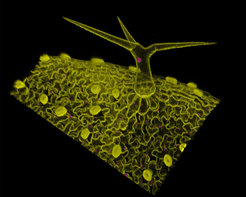
Figure 1. Microscopy image showing the arrangement of different cell types in the Arabidopsis leaf epidermis. Cell outlines are shown in yellow (MYR-YFP marker), and cell nuclei are shown in magenta (H2B-RFP marker). In this image, stomata cells appear scattered throughout the epidermis (small, yellow, ellipsoid-shaped cell pairs), and a large trichome emerging from the epidermis is visible. The undulating cells are pavement cells. In our group, we are studying the nature of the dynamic principles that drive the formation of these cellular patterns.
In developing tissues, cells that are initially indistinguishable from one another differentiate and form tissues containing different cell types. The arrangement of these different cell types is referred to as a cellular pattern. We study how complex cellular patterns are formed in different plant tissues as a result of the interplay between cell signalling, cell growth, and cell division.
At the MPIPZ, we are investigating how cellular patterning is generated in different tissues in Arabidopsis thaliana, with a particular focus on the leaf and sepal epidermis. Across the leaf and sepal surface, trichomes (protective hair cells), stomatal guard cells (which allow gas exchange), and giant cells (large cells that have been related to organ curvature control and plant immunity) are distributed throughout the epidermis and are interspersed among undulated pavement cells. We are evaluating how cell growth and division, cell-to-cell interactions, and noise in gene expression affect the patterning process of these different cell types. In the longer term, we want to understand how complex cellular patterns that include all these different cell types arise and are maintained in plant tissues.
Dynamics of developmental transitions
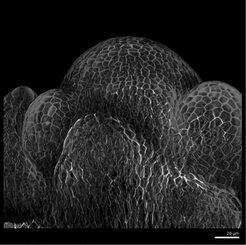
Plants undergo fundamental developmental transitions such as seed germination or the initiation of flowering, known as the floral transition. These processes can be understood as multicellular dynamical systems that undergo important dynamic changes due to their exposure to environmental cues. However, little is known about the dynamics of the key regulators of these complex developmental processes at the single-cell and multicellular levels.
During the floral transition in A. thaliana, the shoot apical meristem (SAM), which is the tissue that produces the aerial organs, undergoes a dramatic morphological change from a flat to a domed structure. In collaboration with the Coupland group at the MPIPZ, we are investigating how the SAM undergoes this change in morphology and how it is influenced by spatio-temporal patterns of gene expression within the SAM. Our studies combine quantitative information at the single-cell level with morphological measures of the SAM. Furthermore, using microscopy data, we are developing the theoretical basis to understand the dynamics of the gene regulatory network underlying the floral transition.
Selected publications
Abley*, K., Formosa-Jordan*, P., Tavares, H., Chan, E. Y. T., Afsharinafar, M., Leyser^, O. and Locke^, J. C.W. (2021) An ABA-GA bistable switch can account for natural variation in the variability of Arabidopsis seed germination time. eLife; 10:e59485. Link
Bertran Garcia de Olalla*, E., Cerise*, M., Rodríguez-Maroto, G., Casanova-Ferrer, P., Vayssières, A., Severing, E., López Sampere, Y., Wang, K., Schäfer, S., Formosa-Jordan, P. and Coupland, G. (2024). Coordination of shoot apical meristem shape and identity by APETALA2 during floral transition in Arabidopsis. Nature Communications, 15(1), 6930. Link
Clark*, F. K., Weissbart*, G., Wang*, X., Harline, K., Li, C., Formosa-Jordan^, P., and Roeder^, A. H. K. (2024). A common pathway controls cell size in the sepal and leaf epidermis leading to a non-random pattern of giant cells. bioRxiv. Link
Formosa-Jordan*, P., Teles*, J. and Jönsson, H. (2018) Single-cell approaches for understanding morphogenesis using Computational Morphodynamics. In: Morris R. (eds) Mathematical Modelling in Plant Biology. Springer, Cham Link
Landrein, B., Formosa-Jordan, P., Malivert, A., Schuster, C., Melnyk, C. W., Yang, W., Turnbull, C., Meyerowitz^, E. M., Locke^, J. C.W. and Jönsson^, H. (2018) Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc. Nat. Acad. Sci. USA. 115:1382-1387. Link
Lakshmi Vadde*, B.V., Russell*, N.J., Bagde, S.R., Saint-Antoine, M., Brownfield, B., Mughal, S., Apprill, L. E., Khosla, A., Clark, F. K., Schwarz, E. M., Alseekh, S., Fernie, A.R., Singh, A., Schrick, K., Fromme, J. C., Skirycz, A., Formosa-Jordan^, P. and Roeder^, A. H.K. (2024) The transcription factor ATML1 maintains giant cell identity by inducing synthesis of its own long-chain fatty acid-containing ligands. bioRxiv. Link
Meyer*, H. M., Teles*, J., Formosa-Jordan*, P., Refahi, Y., San-Bento, R., Ingram, G., Jönsson^, H., Locke^, J. C.W. and Roeder^, A. H.K. (2017) Fluctuations of the transcription factor ATML1 generate the pattern of giant cells in the Arabidopsis sepal. eLife, 10.7554/eLife.19131. Link
Russell, N.J., Belato, P.B., Oliver, L.S., Chakraborty, Roeder, A. H.K., Fox, D.T., Formosa-Jordan, P. (2025) Spatial ploidy inference using quantitative imaging. bioRxiv. Link
(*: equal contributors; ^: co-corresponding authors)
To see the full list of publications, see the following Link

