Marques: Genomic differentiation in holocentric carnivorous Drosera plants
This project will be supervised by André Marques at the Max Planck Institute for Plant Breeding Research
Abstract:
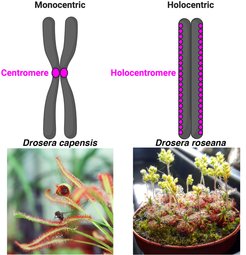
Most studied and sequenced organisms have one single centromere in each chromosome, i.e. monocentric chromosomes, visible as a primary constriction during metaphase. However, several animal and plant species have evolved holocentric chromosomes, where each chromosome harbors hundreds of small centromeres distributed along their entire lengths. Thus, both genome and cell-division related adaptations are expected to be found in such holocentromere-based genomes. This project aims to address how transition from monocentric to holocentric chromosome has influenced genome organization and meiotic recombination using the carnivorous plant genus Drosera as a model system. The genus Drosera is a very exciting group to study adaptation to holocentricity because closely related monocentric and holocentric species can be found. The selected PhD student will be mainly involved in bioinformatic analyses, including genome assemblies, scaffolding and construction of recombination maps, thus, good bioinformatic experience is desired. However, upon interest of the candidate wet-lab experiments could be also undertaken and discussed. Thus, both wet-lab and bioinformatics students may apply for this project. This study will provide a deep knowledge about the impact of transition to holocentricity on genome organization and meiotic recombination dynamics.
Group homepage: https://www.mpipz.mpg.de/marques
Key publication: Hofstatter PG, Thangavel G, Lux T, Neumann P, Vondrak T, Novak P, Zhang M, Costa L, Castellani M, Scott A, Toegelová H, Fuchs J, Mata-Sucre Y, Dias Y, Vanzela ALL, Huettel B, Almeida CCS, Šimková H, Souza G, Pedrosa-Harand A, Macas J, Mayer KFX, Houben A, Marques A. Repeat-based holocentromeres influence genome architecture and karyotype evolution. Cell. 2022 Aug 18;185(17):3153-3168.e18. doi: https://doi.org/10.1016/j.cell.2022.06.045