Evolution of Meiosis
Centromeres are functional DNA sequences that link together pairs of sister chromatids, identical chromosome copies that are formed by DNA replication.
Based on centromere organization, chromosomes are essentially classified into two main types:
- monocentric chromosomes – one centromere domain per chromosome
- holocentric chromosomes – multiple centromere domains distributed genome-wide.
While monocentric chromosomes are regarded as the standard mode of centromere organization, holocentricity has evolved several times independently in animals and plants, on each occasion with specific adaptations. Holocentric plants have evolved several specific adaptations during meiosis, e.g., chiasmatic and achiasmatic inverted meiosis, which postpone homolog segregation to the second meiosis. Thus, holocentric plants do not only offer an exciting model in which to study how these adaptations emerge during evolution but are also of interest for comparative biology. In our team, we aim to decipher the molecular mechanisms underlying the meiotic adaptations observed in holocentric plants.
Most of what we know regarding how holocentric organisms adapt during meiosis is derived from the animal model Caenorhabditis elegans. A holocentric plant model is necessary, because this kind of chromosomal organization evolved independently in different lineages, and thus all adaptations to deal with it are also expected to be different. In our group, we aim to understand the impact of this unique chromosome structure on the genome evolution and meiotic adaptations of holocentric plants from the genus Rhynchospora (Cyperaceae) in which holocentricity is regarded as a shared feature.
GENOME ORGANISATION AND EVOLUTION OF HOLOCENTRIC PLANTS
With the recent advances in long-read sequencing technologies, high-quality genome assemblies of non-model species have become feasible. We have sequenced the genomes of the break-sedge Rynchospora pubera (2n=10) and related Rhynchospora species in order to establish holocentric plant models with diverse genome features. Analysis of the genomes of these holocentric plants is allowing us to further our understanding of how holocentricity evolved in land plants. Some of the specific questions that we are interested in are, how does chromosome structure influence genome evolution in this highly diverse group? Does meiotic recombination occur in or around centromeric regions?
MEIOTIC RECOMBINATION AND INVERTED MEIOSIS IN HOLOCENTRIC PLANTS
Monocentric organisms show restricted meiotic or even non-meiotic recombination at and near centromeres (cold regions), but the reasons for this remain unclear. We therefore aim to better understand how meiotic recombination is regulated in plants with holocentric chromosomes. Using cutting-edge technologies, we are performing analyses with the aim of characterizing meiotic recombination rates and identifying the meiotic proteins involved in the evolution of meiotic adaptations in these organisms. Using holocentric plants as a model to understand how meiotic recombination is regulated at centromeric regions should unveil new strategies to interfere with recombination in monocentric organisms.
INVERTED MEIOSIS
Holocentric plants engage in an unusual mode of meiotic division called inverted meiosis, characterized by the early segregation of sister chromatids at the end of first meiotic division. In the genus Rhynchospora, both chiasmatic (Rynchospora pubera) and achiasmatic (Rynchospora tenuis) inverted meiosis are observed. Genes related to cohesion, condensation, and chiasmata may play important roles in the structural changes associated with inverted meiosis, and special focus will be placed on studying the role of these genes.
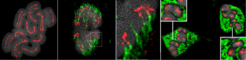
From left to right: Rhynchospora pubera, 1- Mitotic chromosomes showing line-like holocentromeres, 2- meiotic anaphase I showing segregation of sister chromatids with restructured cluster-centromeres, 3- zoomed view of 2, 4- meiotic metaphase II showing the postponed segregation of homologous chromatids. Chromatids (grey), CENH3 (red), tubulin (green).
