New paper on the phylogeny of the Brassicaceae
A recent study from the Max Planck Institute for Plant Breeding Research in Cologne, published in the New Phytologist, helps resolve these issues by reporting new insights into the relationships among Brassicaceae species
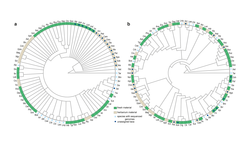
The mustard family Brassicaceae (also known as Crucifers, from the cross-like form of their flowers) comprises ca. 4000 species, including economically important crops, such as cabbage and canola, many species adapted to extreme environments, noxious weeds, and the model plant Arabidopsis thaliana. Despite their importance, the relationships among major lineages in the family have remained unresolved. This gap in our knowledge is an impediment for understanding the sequence of genetic events that led to trait diversity and the many ecological adaptations seen in this family of plants. Lack of such information can also slow down efforts towards sustainable crop improvement through transferring traits from one species into another by conventional or new breeding technologies.
A recent study from the Max Planck Institute for Plant Breeding Research in Cologne, published in the New Phytologist, helps resolve these issues by reporting new insights into the relationships among Brassicaceae species. The study involved close international collaboration and the first author of the paper, Alexander von Humboldt post-doctoral Fellow Lachezar Nikolov worked under the supervision of the Director Miltos Tsiantis and together with Philip Shushkov (California Institute of Technology), population geneticists Dmitry Filatov and Bruno Nevado (University of Oxford), the expert on crucifer morphology and taxonomy Ihsan Al-Shehbaz (Missouri Botanical Garden) and the plant evolutionary biologist Donovan Bailey (New Mexico State University). Nikolov et al. inferred the relationships of 79 Brassicaceae species, which represent 50 of the 52 currently recognized main lineages (known as tribes) in the family, using several hundred nuclear genes spread throughout the genome. A third of the samples were derived from herbarium material, some of the samples dated back to the 19th century, which facilitated broad taxonomic coverage of the family and allowed to study taxa that are difficult to obtain from cultivation or by collecting in the wild. The study found that the diversity of the mustard family falls within six major lineages and provided support for novel relationships among tribes. The authors also resolved the position of 16 taxa that have not been previously assigned to a tribe. Such broad phylogenetic studies, which clarify the relationships among species, provide an essential comparative framework for framing knowledge derived from well-studied model systems in the context of broad diversity seen in nature. Nikolov et al. then went on to provide an example of how their phylogeny can be used to investigate trait evolution by studying leaf shape diversity in the mustards. They identified genome‐wide expression signatures that distinguish simple from complex leaves, found new candidate genes for leaf shape diversification, and identified specific mustard lineages where leaf diversity is particularly pronounced and where more detailed studies should help uncover the precise basis for morphological transitions. The framework provided by this work can now be utilized to study other traits and will enable future efforts to understand the genetic basis for trait diversity in an important group of plants.
