Telomere dynamics in plant meristems and their impact on epigenetic gene repression
Supervision: The project will be supervised by Franziska Turck at the Max Planck Institute for Plant Breeding Research.
Abstract: Telomeres consist of hundreds to several thousand direct repeats of 6-8 base pair sequence motifs. These motifs are bound by protein complexes that protect linear chromosome ends from degradation, chromosome fusion and telomere invasion into other locations. Due to the end-replication-problem, telomeres shorten with every mitotic cycle, unless a specialized eukaryotic reverse-transcriptase, the telomerase, counteracts this process. In animals, telomere shortening is associated with cell-lineage aging and increased telomerase activity is a marker for malignant tumours. In plants, we have recently shown that TELOMERE BINDING FACTORS (TRBs) not only associate with telomeres but also bind to single extratelomeric repeat motifs. TRBs can recruit Polycomb Repressive Complex 2, thereby maintaining the epigenetic repression of target genes. The pathway in particular represses genes with a role in floral development. In contrast, TRBs promote the expression of energy pathway genes, such as genes connected to the Calvin cycle and protein translation (see Figure below). In your PhD, you will explore a possible connection between cell division, telomere-length and epigenetic gene repression. You will establish a method to measure telomere length at individual chromosomes in single cells or small cell populations and then compare telomere length, transcriptomes and epigenetic profiles in meristematic tissues using Fluorescent Marker Assisted Cell Sorting based on transgenic plants expressing fluorescent meristem markers in wildtype and mutant backgrounds.
Link to the Turck group homepage: https://www.mpipz.mpg.de/turck
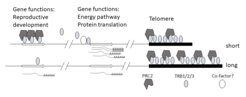
Model of coordination between epigenetic repression, energy pathway regulation and telomeres. Three TRB family proteins bind telomeric repeats and contribute to protect telomeres from degradation. PRC2 may be recruited to telomeres through direct interaction with TRBs, although the function of PcG at telomeres is controversial. Binding of TRBs at the transcription start site of energy pathway genes contributes to maintaining high expression level at these targets. In contrast, TRB binding across H3K27me3 target regions participates in epigenetic repression though PRC2 recruitment. Floral genes are particularly dependent on TRBs for repression. Individual cells can show variation in telomere length, i.e. because their cell lineage has undergone a variant number of cell divisions. It is possible, that competition between TRB binding regions contributes to a coordination of epigenetically controlled developmental processes and metabolism.
