A genetic barcode for the epigenetic repression of the floral program
Plant cells silence their flowering genes with the help of a small DNA-binding protein family which also stabilizes telomeres and promotes the expression of photosynthetic genes. A genetic barcodes shows this family what has to be done. These proteins are thus important mediators of epigenetic imprinting and determine cell fate in plants (Nature Genetics, doi: 10.1038/s41588-018-0109-9).
Multicellular organisms can only exist if each cell knows what to do. Genes that are not required for the job at hand are marked and set aside in tightly packaged chromatin stacks. Some genes are shut down forever, others only temporarily. The term epigenetic has been coined for this type of regulation that seals a cell’s fate. For example, cells in the shoot never resort to the genetic program for roots, but over time need the program for the formation of flowers to produce seeds. Franziska Turck from the Max Planck Institute for Plant Breeding Research in Cologne and her colleagues have investigated how plant cells direct chromatin packaging to the right place and how this may ensure that genes needed for a concerted task, such as flowering, can be mobilized at the same time if necessary.
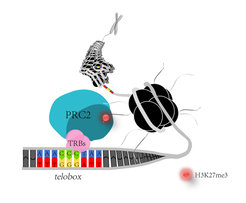
Polycomb repressive complex 2 (PRC2) catalyses the methylation of a specific histone residue (H3K27me3), which serves as epigenetic mark for chromatin compaction. TRB proteins interact with subunits of Arabidopsis PRC2. TRBs also bind a nucleotide sequence motif that is related to plant telomere repeats (telobox). Through this interaction with both, teloboxes and PRC2, TRBs control epigenetic repression of genes involved in floral development.
"We know which proteins determine cell fate by compacting chromatin," says Turck. "An important group are, for example, Polycomb Group proteins. These attach methyl groups as labels to chromatin, a hallmark of epigenetic gene regulation. However, Polycomb Group proteins themselves do not recognize DNA, which raises the question of how they are recruited for this task, "the group leader continues. "We suspect that Polycomb Group proteins gain access to the DNA by piggybacking various DNA-binding proteins. We have now discovered which DNA-binding proteins Arabidopsis plants use to recruit Polycomb Group proteins to secure the epigenetic repression of their floral fate controlling genes. "
For this purpose, Turck and her colleagues have used the genetic model plant Arabidopsis to search for mutants that have a phenotype very similar to plants whose Polycomb Group genes are no longer active. In such a case, it is likely that the mutant proteins are part of the same regulatory pathway. Polycomb Group mutant plants can be recognized by the fact that they are unable to control the timing of developmental processes. Turck and colleagues have selected Arabidopsis mutants that turn on the floral organ-forming program as soon as they germinate, not just when the shoot has the necessary maturity. As consequence, leaves are visibly misshapen at a very early seedling stage.
Further analysis showed that the corresponding Arabidopsis mutants possessed defective TRB proteins. TRB stands for Telomere Repeat Binding proteins and refers to a small family of related proteins for which two apparently very different functional roles had already been described. TRB proteins bind to and stabilize the so-called telomeres at the ends of chromosomes, thereby contributing to the integrity of chromosome and thus of the entire cell. Furthermore, TRBs cause certain genes to be expressed at higher levels. These tasks are dependent on the recognition of a defined DNA sequence, a so-called recognition motif, by TRBs. If this recognition motif is loosely distributed over target genes, a particular barcode is created, which recruits Polycomb Group proteins through a direct interaction with TRB proteins leading to stable gene repression. If TRB proteins are involved in promoting gene expression or telomere stability, another arrangement of the recognition motif is used that does not link to Polycomb Group protein recruitment. How exactly these motifs based barcodes work to direct different processes, Turck and her colleagues will try to figure out next.
