Resistance pathway dynamics in plant immunity
Research in the Parker lab
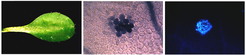
We study how plants activate and control their innate immune responses. Tight regulation of immunity pathways is crucial for combating diseases caused by pathogens, accommodating neutral or beneficial microbes, and prioritizing responses to competing environmental stresses such as drought or nutrition shortage. We analyze plant-microbe interactions and stress network architectures mainly in the model species Arabidopsis thaliana. This provides a molecular genetic and biochemical framework for comparative studies with other (dicot and monocot) plant species to identify fundamentally conserved or clade-specific immunity mechanisms. Our aim is to identify and harness key players and processes in cells and tissues that enable plants to respond effectively to attacking microbes while maintaining growth and physiological fitness. We use approaches ranging from genetics, CRISPR-Cas9 technology, RNA-seq, ChIP-seq, LC-MS/MS mass-spectrometry, live-cell imaging/FRET-FLIM and protein biochemistry/structure-function analyses to computational phylogenomic and evolutionary co-occurrence studies.
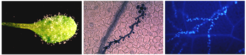
Current Projects in the lab are:
1. NLR immunity - decision-making and execution.
Plant intracellular NLRs mediate effector-triggered immunity (ETI), working as pathogen-activated sensors (receptors) or host-activated immune signalling (helper) components. Major insights to NLR receptor activation mechanisms were made in recent years but how activated NLRs then mobilize anti-microbial defences remained unclear. Using Arabidopsis and N. benthamiana we’ve been characterizing processes that connect NLRs to transcriptional defences and regulated cell death (RCD).
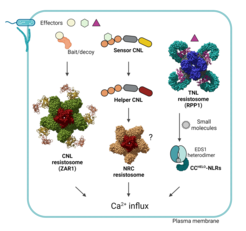
In collaboration with the renowned protein structural chemist Prof. Jijie Chai (now at Westlake University, Hangzhou ), we determined the biochemical mechanism of plant Toll-Interleukin1-Receptor (TIR) TIR-domain NLR (TNL) activation. Pathogen-activated TNL receptor oligomers (resistosomes), and other immune-responsive TIR-domain proteins, catalyze the production of a set of ribosylated nucleotides which specifically activate EDS1-helper NLR immunity branches. We’re examining EDS1-helper NLR subcellular sites and modes of action for mediating host calcium-dependent nuclear transcription and localized cell death.
2. Biotic stress network architectures across plant lineages.
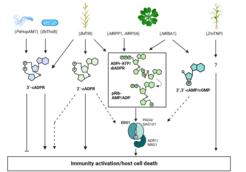
Using known Arabidopsis immune pathway properties as a reference, we’re investigating the defence network architectures of other plant species such as the solanaceous model Nicotiana benthamiana and monocot crops, barley and rice. This analysis is starting to reveal conserved working principles but also interesting clade-specific variation in the usage of defence signalling modules and pathways. On one hand, we’re exploring the evolutionary origins and catalytic activities of different TIR-domain containing proteins. TIR-catalytic versatility in generating bioactive nucleotide-based SMs across kingdoms is an exciting new area of immunometabolism. On the other hand, with Thorsten Nürnberger & colleagues (ZMBP, Tübingen) and Haitao Cui (Fujian Agricultural University) we’re characterizing Arabidopsis (or Brassicaceae)-specific processes in tissue-level disease resistance potentiation.
3. Tracking and engineering immune-stimulating nucleotides.
Together with the organic chemistry group of Stephanie Kath-Schorr at University of Cologne, we’re using the newly discovered plant ribosylated nucleotide molecules as a platform to design and synthesize modified immune-stimulating nucleotides. Strategies are being tested for the delivery and release of bioactive SMs in plant cells, and measuring immunity outputs. By exploiting structurally characterized SM-receptor and signalling variants we aim to track activities of natural and modified SMs in dicot and monocot plants as a path towards designing an immune-potentiating agent for crops.
4. Defence metabolites impacting plant interactions with beneficial and pathogenic fungi
Here we are exploiting genetic material in Arabidopsis to investigate how plants discriminate between beneficial, neutral and harmful (pathogenic) fungi associating with roots or leaves. We’re examining a root accommodation programme between Arabidopsis and a natural fungal endophyte which the host is normally able to contain and benefit from in terms of resilience to biotic or abiotic stress. We’re testing the effects of different immunity mutants and nutrient provision on plant-endophyte colonization outcomes. In focussed assays, we’re studying the interplay of host plants (leaves and roots) with pathogenic or beneficial fungi in modulating coumarin metabolism and how this affects plant health. Coumarins are made from phenylpropanoid precursors and occur broadly across seed plants. Depending on the coumarin chemical structure, it can possess iron-chelating or non-chelating activity, both of which have potential to shape plant – microbe interactions.
If you’re interested in joining the group to pursue research in any of these areas, please contact Jane Parker with a CV and a short description of your scientific interests.



