New insight into leaf shape diversity
Pleiotropy influences the genes shaping leaf diversity
Many of us probably remember the punnett squares by which we were introduced to the idea of genetic inheritance in school: a dominant allele in each of my brown-eyed parents hides a recessive allele that explains my blue eyes. Most genetic inheritance, of course, is much more complex. Differences in how we look are usually produced not by a single gene, but rather by interactions between several genes, across a gene regulatory network. Conversely, a single gene can affect not just one but many aspects of an organism’s development – and these are often unrelated. This second phenomenon, which scientists call “pleiotropy” from the Greek word for “many ways”, is central to one of the more interesting puzzles of evolutionary biology. A paper published in Genes and Development sheds some light on this puzzle.
Miltos Tsiantis, a director at the Max Planck Institute for Plant Breeding Research, explains pleiotropy like this: "Suppose that a bird developed an evolutionary modification to the shape of its wing, and this change made flying easier, or faster. But if the gene that modifies the wing shape also changed the shape of the foot, making it harder to grip branches, or changed something else that made it less attractive to potential mating partners, then that modification to the wing shape is unlikely to be successful, and persist in populations”. The reason that this is an important issue is that many genes do not only influence the development of a single trait – in this case wing shape – but also others that are sometimes completely unrelated. Pleiotropy is thus a potential obstacle to evolutionary change, “so evolution needs to find ways to work around it”, says Tsiantis.
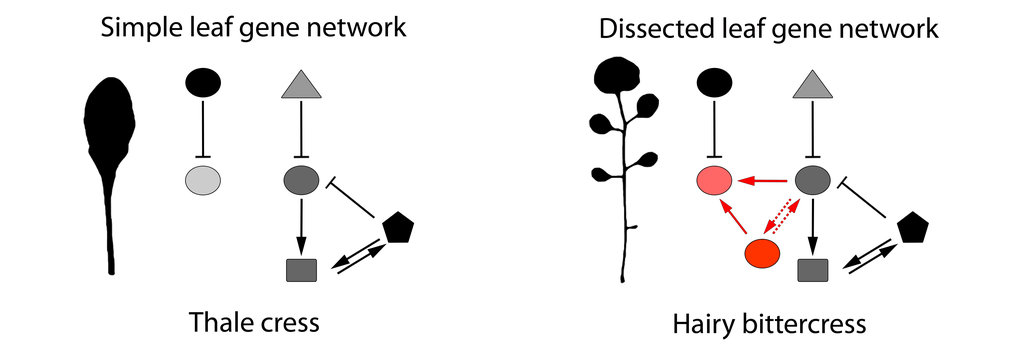
In their current study, Tsiantis and lead author Rast-Somssich examined observable differences between two related plant species, aiming to make sense, at the genetic level, of what is causing those differences: is there a logic to the evolutionary change, and can this logic predict the genetic basis of change? Their experiment focused on differences in leaf shape between thale cress, which has simple, undivided leaves, and hairy bittercress, which has dissected leaves divided into leaflets However, unlike our blue eyes/brown eyes example from school genetics, there is a more complex gene network that determines whether a leaf will have a simple or dissected shape. And the challenge of unravelling this network was part of the fun for Rast-Somssich “For me, it is most exciting to see that cis-regulatory changes of an individual gene can create novel genetic interactions in one species that could not be predicted by what was known in the other.”
One key difference between the leaf network operating in thale cress and hairy bittercress is the action of two genes that control bittercress leaf shape. Of the two genes, one plays a more dominant role and is more pleiotropic than the other. The authors demonstrate that it is this other, less pleiotropic gene, which evolved a greater capacity to contribute to evolutionary change in leaf shape between species. This result supports previous research showing that changes in regulatory sequences of genes can circumvent pleiotropy by allowing redeployment of potent regulators in space and time. “This targeted redeployment allows changes without messing everything up”, says Tsiantis. So the discovery of this tendency indicated some predictability in how things change.
However, the results of this new study also show something that adds a layer of complexity to our understanding of these mechanisms for how evolution works. Very potent developmental regulators may be less able to change and acquire new characteristics, than the less potent alternatives. “At least in this case”, says Tsiantis, “the protein that is less potent seems to have more leeway to evolve more diverse variants.” So this is a second way of circumventing pleiotropy. The less potent regulators may be more likely to produce new linkages in a gene network, connecting the activities of smaller sub-networks that previously did not talk to each other, contributing to a different function in tissue growth.
Effectively, this suggests that diversity is generated not simply by the redeployment of conserved gene functions but also, perhaps at least as much, by the new relationships these genes create within an existing network.